| Title of the project | Establishment of a system for the use of DNA markers for genetic identification and verification of varietal authenticity and purity of major cereals and brassicas as a basis for the quality production of seeds, food and feed |
|
Code / SICRIS |
V4-1806 |
|
Duration |
1. 11. 2018 – 31. 10. 2021 |
|
Head |
|
| Research group | List of researchers and technical staff |
|
Research organisation |
|
|
Financing |
|
|
Bibliography of the project |
|
|
Short description of the research project
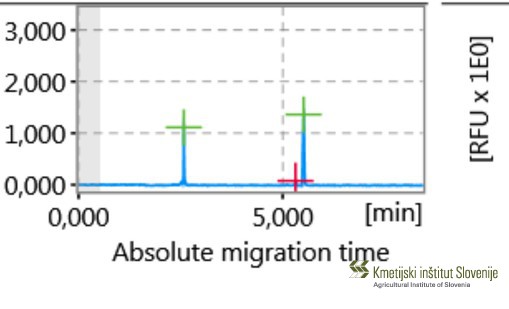
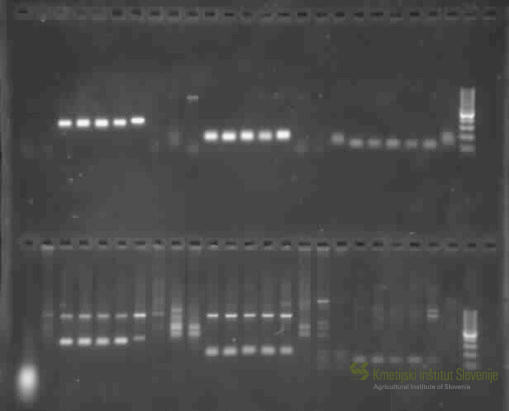
The key components of ensuring food safety and risk reduction for a successful agricultural production are seed quality and varietal genuineness, which are adapted to Slovenian growth conditions. New regulations, established by International Seed Testing Association (ISTA) on January 1st in 2017, are introducing more reliable procedures to verify varietal genuineness based on DNA markers. Giving the relevance of this issue, ISTA is currently in preparation for releasing a DNA-based Variety Identification Manual, which will be published in 2019. In its 2016 – 2017 report, the ISTA working group introduced wheat Short Sequence Repeats (SSR) markers. SSR markers are in general mostly used molecular tool for different plant breeds, although for some breeds more precise Single Nucleotide Polymorphism (SNP) markers are being introduced. (ISTA News Bulletin, 154, October 2017). In cooperation with Organization for Economic Co-operation and Development Seed Schemes (OECD), the Service for Official Certification of Seed and Plant Propagation Material of Agricultural Plants (SUP) at Agricultural Institute of Slovenia was called upon to define biochemical and molecular analysis for varietal purity testing, identification and hybridization analysis within individual plant species, which also needed to be defined. In this regard a lot of attention is being drawn to application of DNA markers, while biochemical markers are increasingly more being replaced by the former. Lately, plant breeders are taking effort for each variety description to also include prescriptive methods for genetic identification and accessible genetic profile of specific loci in genome for effective and reliable identification of varieties. Subsequently plant breeders are interested in introducing new, effective and reliable variety identification methods to protect their “breeders’ rights”, for which DNA markers are most suitable. Most difficulties with official certification of plant material during vegetation occur with plants with atypical traits, especially within cereal and crucifer species, where variety identification is challenging. Same difficulties occur during laboratory analysis of seed quality, where seed morphologic traits can cause ambiguity when tested for varietal purity and germination, causing dubiety in plant variety, species and even genus. New varieties are becoming less indistinguishable by morphological traits, which make visual identification problematic. Within proposed project, we aim to establish a reliable and effective genetic tool which will be used for target identification of genus/species/variety of important cereal and crucifer members, if varietal purity and identification analysis of seed and plant material samples will not be able to provide definite results. Such established molecular marker system could enable genome-specific and variety differentiation both inter- and intra-species of cereal and crucifer species, produced in Slovenia. The established methods will also support with PRP (PRP control), especially regarding GEN_SOR demands. The goals of proposed project are: - Development and establishment of DNA marker-based genetic identification system for genus/species/variety of agronomical important species of cereals and crucifers registered in Slovenian or European variety list and whose plant material is grown or officially certified in Slovenia (e.g. Triticum aestivum L. Emend. Fiori et Paol. (Common wheat); Brassica napus L. (partim.) (Rapeseed); Fagopyrum esculentum (Moench) (Common buckwheat); Hordeum vulgare L. (Barley); etc.). This collection will also include species which are difficult to identify and distinguish on phenotypic and morphological levels of seeds, seedlings and plant traits in seed production. - Optimization of current official certification procedures and verification methods of varietal genuineness, purity and determining detection levels of atypical plants (for self-pollinating hybrid varieties) and other plant varieties in seed tillage and seed partition. Moreover, application of DNA markers represents supporting tool in the case of doubts regarding to variety identification within the official procedure of certification establishing better quality of seeds on the market. - Preparation of methodological documents and information for establishing databases of varieties’ allele profiles registered in List of varieties, grown and officially certified in Slovenia Abstract of the finished project Varietal authenticity and high-quality seed of varieties adapted to Slovenian production conditions are of key importance for successful agricultural production and thus for ensuring food security and reducing risk in agricultural production. Recent regulations of the International Seed Testing Association (ISTA) introduce more reliable methods of authentication of varieties based on the use of DNA markers. Within the framework of the OECD (Organization for Economic Cooperation and Development) Seed Systems Working Group, the Service for Official Certification of Seeds and Seedlings of Agricultural Plants (SUP) at the Agricultural Institute of Slovenia, as the Slovenian certification body, was asked to comment on the introduction of biochemical and molecular analyzes to verify varietal purity, identification and hybridization within individual plant species for which analyzes can be performed. Also from this side, the application of DNA markers is strongly emphasized, while the analyzes based on biochemical markers are abandoned. Recently, breeders have increasingly sought to ensure that each variety has a method of genetic identification and an accessible genetic profile of specific loci in the genome that can be used to reliably and efficiently distinguish varieties from one another. Consequently, breeders are increasingly turning to new reliable methods of effectively distinguishing varieties from one another through the use of DNA markers. Indeed, the certification of seeds during the growing season also raises the problem of identifying varieties of atypical plants, especially in cereal and cruciferous species. The usefulness of genetic identification is shown during laboratory analysis of seed quality, where seed morphologic traits can cause ambiguity when tested for varietal purity and germination, causing dubiety in plant variety, species and even genus. Indeed, new varieties are becoming more and more similar morphologically, so that their identification by visible characteristics is becoming more and more difficult. For the purposes of the project activities, we first prepared lists of varieties included in the CRP after verifying the valid SLO list of varieties and seed production through official standard samples. A total of 54 cereal and cruceferous varieties were included in the project. In the analyzes conducted at the ISTA-accredited seed laboratory KIS, we performed seed quality analysis (determination of purity and germination) for each variety according to the valid ISTA rules. Based on seed morphology, we formed subgroups if seeds of the same variety differed morphologically. The seeds and seedlings of each variety were also photographed. After the analyzes in the seed testing laboratory, the seedlings were taken to the genetic laboratory for analysis with DNA markers. Each of the 54 cultivars was represented by 4 (for self-pollinating plant species) or 8 (for out-pollinating plant species) individual plants during genotyping, so we were also able to obtain information on intra-varietal variability and allele frequencies. We had a total of 277 samples for genetic analysis. We then optimized the DNA extraction method for each plant species individually. Six different DNA extraction protocols were tested, and three different protocols were used depending on the species (extraction with three commercial kits and/or with an automated nucleic acid extraction system). In addition, for genome-specific and varietal segregation within/between cereal and cruciferous species, 46 different DNA markers were identified based on databases and scientific articles, including 3 different marker systems: 43 SSR (Simple Seguence Repeat) markers, 2 SCAR (Sequence Characterized Amplified Region) markers, and one CAPS (Cleaved Amplified Polymorphic Sequences) marker, respectively. Thirteen DNA markers were used for differentiation within the wheat/rye/triticale/spelt complex; for differentiation within the genus Avena, 7 DNA markers; for differentiation within the genus Hordeum, 6 DNA markers; for diversity and differentiation within and between common and tartary buckwheat, 11 DNA markers; for diversity and differentiation within and between species from the family Brassicaceae, 9 DNA markers. For each marker, reaction mixtures and amplification conditions were optimized for polymerase chain reaction (PCR) and fragment analysis to obtain a qualitative (presence/absence of propagation at each locus) and quantitative (allele) profile of the variety. The results of the fragment analysis with the allele profiles of the cultivars in the form of codominant matrices required additional processing with statistical programs and population genetics tools to obtain information on allele frequencies and intra-specific diversity within each cultivar. These results will be part of a comprehensive methodological document that we are still preparing. As part of the project, we have also empirically determined the limits of detection for the smallest possible amount of plant tissue detectable with standard PCR. It actually represents the presence of atypical plants in the bulk sample/crop and we are able to determine them at a qualitative level. We have found that at the qualitative level we can detect even very small differences in the genome for a single marker, if any. For each of the specific SSR markers we determined to successfully discriminate between parents and hybrids at a 1% of atypical plants using agarose gel electrophoresis at the qualitative detection level. If we want to determine the differences at the lower proportions of atypical plants (i.e. 0.1% and/or 0.3% according to the scheme for different categories of certified seeds), we should continue the analysis at the level of allele detection (more expensive, time-consuming and methodologically demanding). However, accurately determine the differences between genotypes on a base pair, which is optimal and absolutely reliable. Based on the quantitative results, we have also drawn a phylogenetic tree, which shows that we are also able to distinguish the proportions of atypical plants that are less than 1%, but with lower reliability at the qualitative level. The main objective of the project was the successful implementation of a system for reliable identification of cereals and cruciferous species using DNA markers, which now allows fast, efficient and financially acceptable verification of varietal authenticity and purity as part of the official certification of cereals and cruciferous species. Established protocols and variety- or species-specific DNA markers can also be used to determine the authenticity of local varieties and to control food safety when comparing field crops with those from storage or in flours. In fact, the methods of the project were also validated on actual samples apperaing at KIS/SUP to solwe real certification problems/doubts using genetics for the following sets: common and naked oats; wheat, spelt and triticale; B. rapa and B. napus; distingushing among barley varieties; distingushing among common buckwheat varieties. We have also prepared a basic/overarching document at the national level, which methodically defines the procedures for identification of cereal and crucifer varieties and databases with genetic profiles of varieties/alleles registered in the Variety List in Slovenia or whose material is officially certificied. This document will be available in printed form to the public and all interested parties by summer 2022. Based on the data in this document, the KIS genetics laboratory will now be able to quickly and reliably perform genetic analyzes that effectively resolve doubts about variety identity and purity for the needs of the certification body (in accordance with OECD recommendations), as well as the needs of the ISTA-accredited laboratory, the inspection service, UVVHR, MKGP, farmers, seed importers and also breeders themselves. Publikacija: Metode, postopki in rezultati preverjanja sortne pristnosti žit in križnic (.pdf) |
|